|
Pangolin
|
Contains a linear ozone chemistry scheme. Requires HDF5. More...
Public Member Functions | |
| subroutine | init_chemistry (grid) |
| subroutine | set_global_position (grid) |
| Set the global index of the first grid cell. More... | |
| subroutine | read_chemistry () |
| subroutine | write_coef_debug (month, fname, nb_lat2) |
| subroutine | chemistry_iteration (grid, k) |
| One chemistry step for a grid. More... | |
| double precision function | linear_interpol_coef (i, day, month, year, offset) |
| Suppose the coefficients are given the 15 of each month. Interpolate in time at a given day. More... | |
| subroutine | linear_interpol_all_coef (A, day, month, year, offset) |
| Interpolate coefficients linearly in time. Assume the coefficients are independant from longitude. More... | |
| double precision function | ozone_scheme (q_cur, T_air, A) |
| No heterogenous chemistry and no ozone colomn terms. More... | |
| subroutine | check_hdf5_error (error, msg) |
| integer function | get_month (iter) |
| Returns the current month to get the proper coefficients. More... | |
| subroutine | check_file_size (dataspace, nb_lat2) |
| subroutine | free_chemistry () |
Public Attributes | |
| integer | nb_months = 12 |
| integer | nb_coefs = 9 |
| 9 coefficients More... | |
| integer | month = -1 |
| double precision, dimension(:,:,:), allocatable | a_i |
| Array of A_i, where A_i is of the form (nb_months, nb_lat) More... | |
| integer | start = -1 |
| First cells in the latitude array for the current process. More... | |
| character(*), parameter | fname_chem = "chemistry.F90" |
| character(2), dimension(9), parameter | list_coefs = (/ "A1", "A2", "A3", "A4", "A5", "A6", "A7", "A8", "A9"/) |
| double precision | dt_chem = -1 |
Contains a linear ozone chemistry scheme. Requires HDF5.
The ozone tendency is equal to:
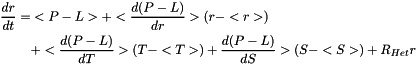
with
Terms between < > denote two-dimensional variables obtained at equilibrium from the MOBIDIC 2D photochemical model. Other terms are three-dimensional variables generated by the 3D Circulation Model.
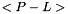 (units : s-1)
(units : s-1)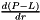 (units : s-1)
(units : s-1)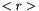 (units : none (volume mixing ratio))
(units : none (volume mixing ratio))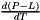 (units : s-1 K-1)
(units : s-1 K-1)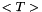 (units : K)
(units : K)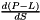 (units : cm2 molecule-1 s-1)
(units : cm2 molecule-1 s-1)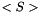 (units : molecule cm-2)
(units : molecule cm-2)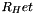 (units : s-1 (to be used only if T<195K during daytime at high latitude))
(units : s-1 (to be used only if T<195K during daytime at high latitude))This parameterization is described by Cariolle et al., 1986 (version 1) and the present version 2 by Cariolle and Teyssedre (2007). Version 2 has been improved over version 1 with an update version of the MOBIDIC model. Major changes concern:
The major improvements of version 2 over the version 1 are a reduction of a negative bias in the ozone content in the middle stratosphere (around 10-30 hPa) at low and mid latitudes (compared to HALOE and SAGE data), a reduction f a positive bias in the ozone content in the troposphere and a total ozone content in very good agreement with he TOMS data all year around.
In addition, the version 2 can be used with a cold tracer formulation for calculation of the heterogeneous chemistry (see Cariolle and Teyssedre, 2007). The coefficient A9 gives the cold tracer decay rate (Units: s-1).
The users of the parameterization are requested to give reference to Cariolle and al., 1986 and/or Cariolle and Teyssedre (2007) in any publication of results obtained using this paramaterization, and to refer to the authors in case of problems encountered in implementation or use.
For further informations, you can contact :
Daniel Cariolle
CERFACS - METEO-FRANCE phone: 33 (0)5 61 19 31 71 42, avenue G. Coriolis fax: 33 (0)5 61 19 30 00 F-31057 Toulouse Cedex e-mail: daniel.cariolle@cerfacs.fr
Reference : Cariolle, D. and M. Deque. Southern hemisphere medium-scale waves and total ozone disturbances in a spectral general circulation model. J. Geophys. Res., vol. 91, pp 10825-10846, 1986.
Cariolle, D. and H. Teyssedre. A revised linear ozone photochemistry parameterization for use in transport and general circulation models: multi-annual simulations. Atmos. Chem. Phys., 7, 2183Ã2196, 2007.
| subroutine chemistry::check_file_size | ( | integer(hid_t) | dataspace, |
| integer | nb_lat2 | ||
| ) |

| subroutine chemistry::check_hdf5_error | ( | integer, intent(in) | error, |
| character(*), intent(in) | msg | ||
| ) |

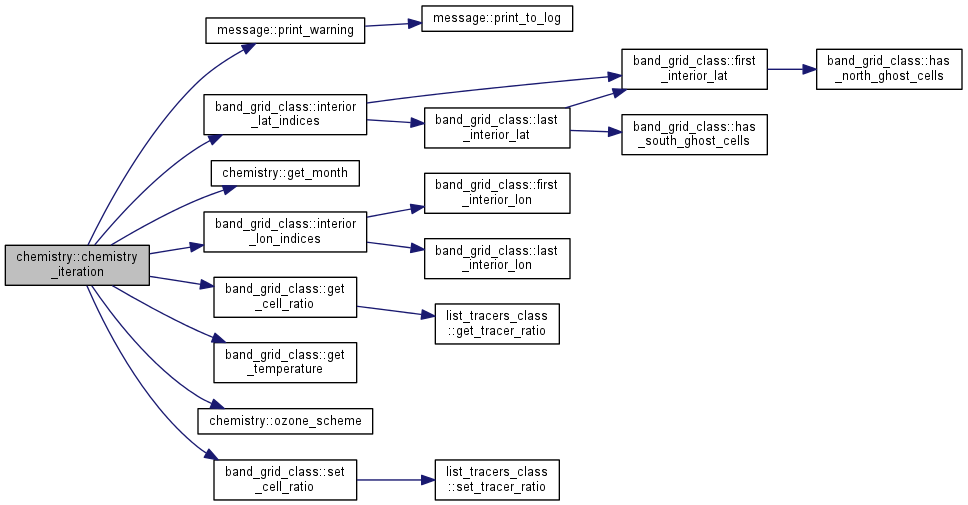
| subroutine chemistry::chemistry_iteration | ( | type(band_grid) | grid, |
| integer, intent(in) | k | ||
| ) |
One chemistry step for a grid.
| k | : current iteration |
| subroutine chemistry::free_chemistry | ( | ) |
| integer function chemistry::get_month | ( | integer | iter | ) |
Returns the current month to get the proper coefficients.
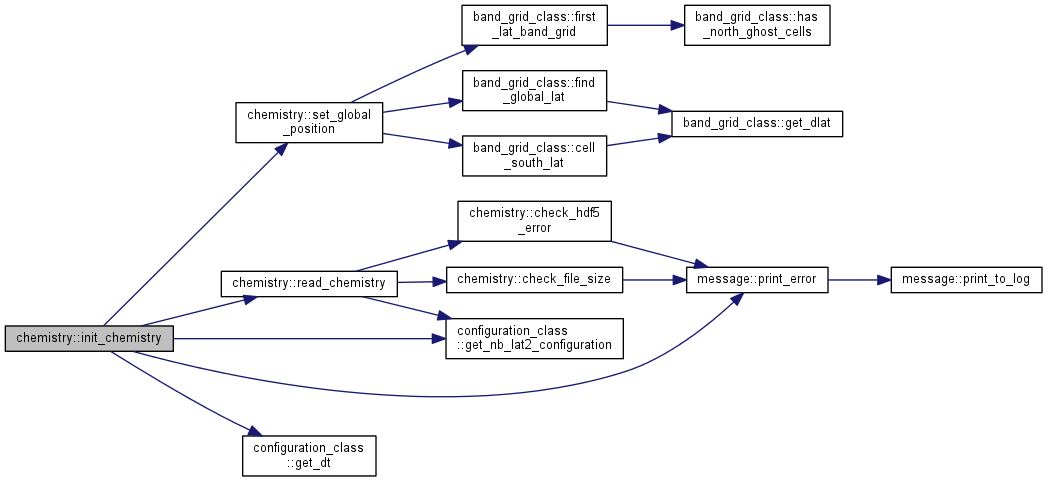
| subroutine chemistry::init_chemistry | ( | type (band_grid) | grid | ) |
| subroutine chemistry::linear_interpol_all_coef | ( | double precision, dimension(5) | A, |
| integer, intent(in) | day, | ||
| integer, intent(in) | month, | ||
| integer, intent(in) | year, | ||
| integer, intent(in) | offset | ||
| ) |
Interpolate coefficients linearly in time. Assume the coefficients are independant from longitude.

| double precision function chemistry::linear_interpol_coef | ( | integer, intent(in) | i, |
| integer, intent(in) | day, | ||
| integer, intent(in) | month, | ||
| integer, intent(in) | year, | ||
| integer, intent(in) | offset | ||
| ) |
Suppose the coefficients are given the 15 of each month. Interpolate in time at a given day.
| day,month | current date |
| i | : coefficient number |
| offset | : offset from start, the first global position |
| double precision function chemistry::ozone_scheme | ( | double precision, intent(in) | q_cur, |
| double precision, intent(in) | T_air, | ||
| double precision, dimension(5), intent(in) | A | ||
| ) |
No heterogenous chemistry and no ozone colomn terms.
| q_cur | : ozone ratio at previous iteration |
| T_air | : temperature at previous iteration |
| q_next | : ozone ratio at next iteration |
| A | : all five coefficients interpolated beforehand |
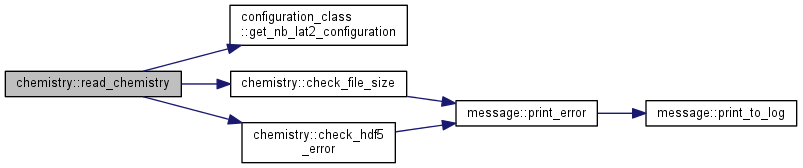
| subroutine chemistry::read_chemistry | ( | ) |
| subroutine chemistry::set_global_position | ( | type (band_grid) | grid | ) |
Set the global index of the first grid cell.
| subroutine chemistry::write_coef_debug | ( | integer | month, |
| character(*) | fname, | ||
| integer | nb_lat2 | ||
| ) |
| double precision, dimension(:, :,:), allocatable chemistry::a_i |
Array of A_i, where A_i is of the form (nb_months, nb_lat)
| double precision chemistry::dt_chem = -1 |
| character(*), parameter chemistry::fname_chem = "chemistry.F90" |
| character(2), dimension(9), parameter chemistry::list_coefs = (/ "A1", "A2", "A3", "A4", "A5", "A6", "A7", "A8", "A9"/) |
| integer chemistry::month = -1 |
| integer chemistry::nb_coefs = 9 |
9 coefficients
| integer chemistry::nb_months = 12 |
| integer chemistry::start = -1 |
First cells in the latitude array for the current process.
 1.8.6
1.8.6