Photo Egor Myznik. Research codes often use custom input formats instead of standards (which can be limiting sometimes). What the format gained in freedom, is lost in accessibility: new users can be puzzled and learn sometimes reluctantly, like a new cuneiform alphabet. So we created translators…
An AVBP simulation is composed of several HDF files (mesh, initial solution, solutbound…) and of ASCII files (run.params, mesh.asciibound, mixture_database…). This second type represents quite an important amount of data sorted in a custom format. PyAVBP offers helpers tools to load or edit these files.
Reading as a Python dict
This small code shows how to load the content of AVBP input files as Python dictionaries.
import pyavbp.tools.data_to_dict as data2dict
#Paths to the ASCII files
my_runparam = './run.params'
asciibound_file = '../MESH/my_mesh.asciibound'
species_file = './species_database.dat'
mixture_file = './mixture_database.dat'
#Turn them into dicts
rp_dict = data2dict.main_runparams_to_dict(my_runparam)
bound_dict = data2dict.main_asciibound_to_dict(asciibound_file)
species_dict = data2dict.main_speciesdata_to_dict(species_file)
mixture_dict = data2dict.main_mixturedata_to_dict(mixture_file)
Now, you can use these dicts to access any value you need:
> print(rp_dict.keys())
dict_keys(['RUN-CONTROL', 'INPUT-CONTROL', 'OUTPUT-CONTROL', 'TPF-LAGRANGE::1', 'THICKENED-FLAME-MODEL', 'POSTPROC'])
> print(species_dict['CH3']['species_formation_enthalpy'])
-66911.0
Note that all these functions convert avbp doubles into floats and avbp integers into int.
Editing a run_params as a python dict
If you need to generate automatically different versions of a run.params file (e.g. for a parametric batch of runs), there is a useful object: RunParamsEditor:
r_m = RunParamEditor("./run.params") #Load
r_m["RUN-CONTROL"]["CFL"]= 0.4 # Change
r_m["RUN-CONTROL"]["CFL2"]= 0.4 # Add stupid keyword 1
r_m["RUN-CONTROL"]["CFL3"]= 0.666 # Add stupid keyword 2
del r_m["RUN-CONTROL"]["CFL2"] # remove stupid keyword 1
r_m["POSTPROC.1"]["cut_planar.average"]= "yes"
r_m.dump("./run.params_modified") #Dump
The peculiar POSTPROC block
The run.params file comes with some caveats.
Look at the following POSTPROC Block:
$POSTPROC
postproc.activate = yes
postproc.dirname = ./OUTPUT_POSTPROC/
cut_planar.name = 00_cut_planar_postproc_dummy#
cut_planar.average = no
cut_planar.save.time = 1.00000000d-04
cut_planar.origin = 0.00000000d+00 0.00000000d+00 0.00000000d+00
cut_planar.normal = 0.00000000d+00 1.00000000d+01 0.00000000d+00
cut_planar.name = 01_cut_planar_postproc_dummy
cut_planar.average = no
cut_planar.save.time = 1.00000000d-04
cut_planar.origin = 0.00000000d+00 0.00000000d+00 0.00000000d+00
cut_planar.normal = 1.00000000d+00 0.00000000d+00 0.00000000d+00
$end_POSTPROC
Some keywords are repeated within the same block. You need to take into account the order to now what value goes where. Technically, these keywords are non-hashable and cannot be used as dictionary keys.
The main_runparams_to_dict function and the object RunParamEditor built on it explicitly add the order of the postproc element in the key.
This is why, if you print the object or the dictionary (print(r_p)), it looks like:
┃ ┣ flame_params_table.tabulated_var (str) : phi
┃ ┣ flame_params_table.temperature (float) : 300.0
┃ ┣ flame_params_table.pressure (float) : 101325.0
┃ ┗ flame_params_table (str) : ./flame_params.h5
┣ POSTPROC.main (dict)
┃ ┣ postproc.activate (str) : yes
┃ ┗ postproc.dirname (str) : ./OUTPUT_POSTPROC/
┣ POSTPROC.0 (dict)
┃ ┣ cut_planar.name (str) : 00_cut_planar_postproc_dummy#
┃ ┣ cut_planar.average (str) : no
┃ ┣ cut_planar.save.time (float) : 0.0001
┃ ┣ cut_planar.origin (str) : 0.00000000d+00 0.00000000d+00 0.00000000d...
┃ ┗ cut_planar.normal (str) : 0.00000000d+00 1.00000000d+01 0.00000000d...
┗ POSTPROC.1 (dict)
┃ ┣ cut_planar.name (str) : 01_cut_planar_postproc_dummy
┃ ┣ cut_planar.average (str) : yes
┃ ┣ cut_planar.save.time (float) : 0.0001
┃ ┣ cut_planar.origin (str) : 0.00000000d+00 0.00000000d+00 0.00000000d...
┃ ┗ cut_planar.normal (str) : 1.00000000d+00 0.00000000d+00 0.00000000d...
Rest assured, when you dump the content of the dictionary (with ruparam_dict_as_avbpstr(dict_) or more simply r_p_dump()) , the splitted keys POSTPROC.main, POSTPROC.0, POSTPROC.1 will be merged in a single POSTPROC block in pure AVBP style…
Visualisation
Once you have made a dict out of these files, you can turn them into YAML files. Then you can use another COOP package to get an overview of the way data is structured in your file. This package is Nobvisual:
import nobvisual
with open('mixture.yml','w') as fout:
yaml.dump(mixture_dict, fout)
nobvisual.visual_treefile('mixture.yml')
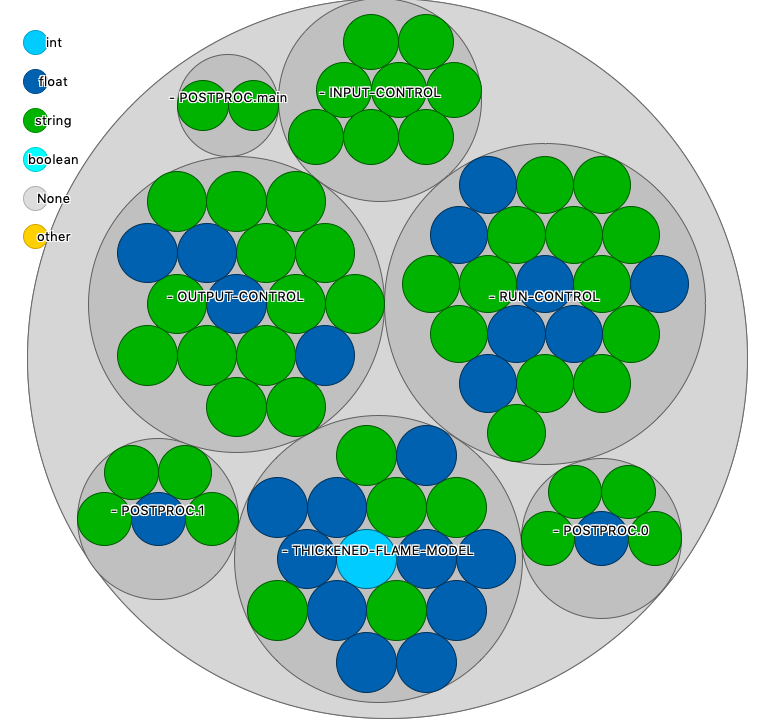
The different blocks of run.paramscan easily be spotted here. The POSTPROC is divided in three parts to make it Hasable as a dictionnary.
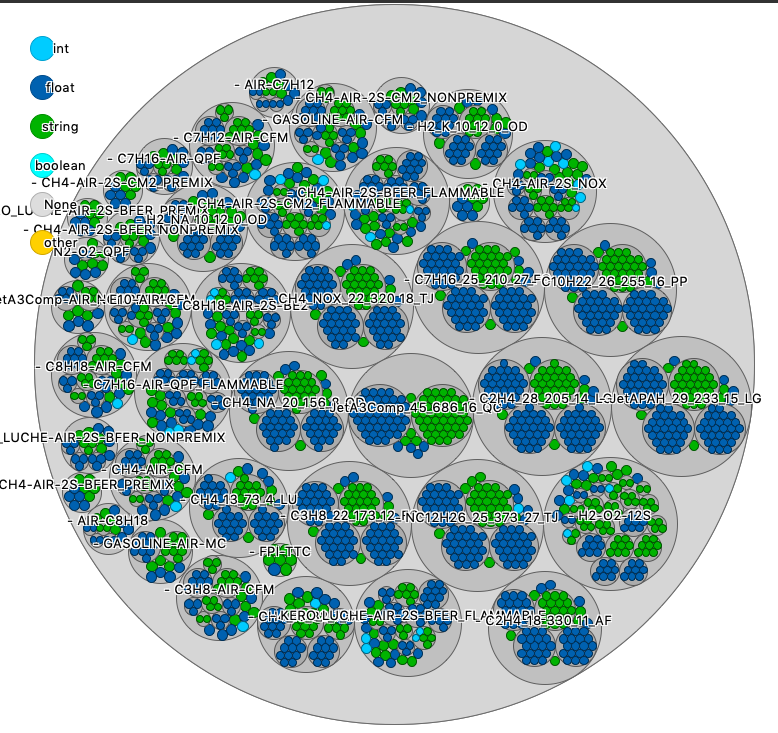
The mixture_database.dat is a large storage file for many physical quantities. You can see all of these datasets, gathered by mixtures.
Takeaway
The in-house parser of AVBP eventually led to custom ASCII files. If the process you are working on need to scale up (be released to an industrial customer for example), feel free to use the PyAVBP helpers tools. These are tested and maintained, not perfect but trying to improve…
