Connectivity filter¶
Description¶
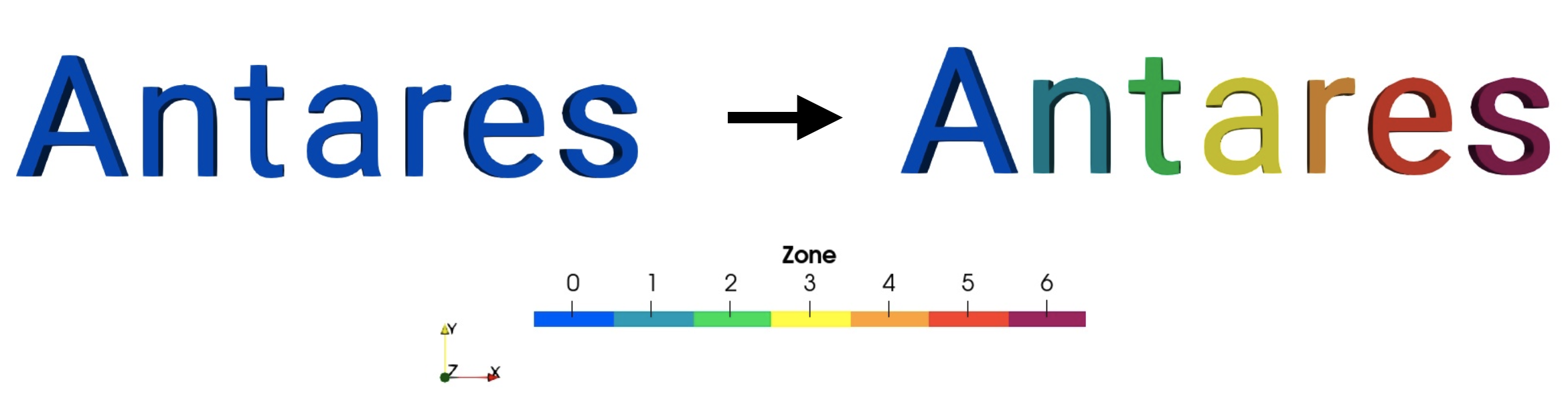
Split a single-zone base into a multi-zone base with the different connected regions.
Parameters¶
- base:
Base The input single-zone base to be split
- base:
- type: str, default=
'node' How cells are considered to be connected to other cells. If
'node', cells are considered as connected if they share at least one node. If'edge', cells are considered connected if they share at least one edge.
- type: str, default=
Preconditions¶
Input base must contain only one zone.
Input base must have a shared geometry
Postconditions¶
The output base contains one zone per each independently connected region in the base.
Example¶
import antares
myt = antares.Treatment('connectivityfilter')
myt['base'] = base
myt['type'] = 'edge'
split_base = myt.execute()
or
import antares
split_base = antares.treatment.ConnectivityFilter(base=base, type='edge')
Example¶
The following example shows how use the treatment
import antares
import os
output_folder = os.path.join("OUTPUT", "TreatmentConnectivityFilter")
os.makedirs(output_folder, exist_ok=True)
reader = antares.Reader('hdf_cgns')
reader['filename'] = os.path.join('..', 'data', 'CONNECTIVITY_FILTER', 'antares.cgns')
base = reader.read()
treatment = antares.Treatment('connectivityfilter')
treatment['base'] = base
smoothed_surface = treatment.execute()
writer = antares.Writer('hdf_antares')
writer['base'] = smoothed_surface
writer['filename'] = os.path.join(output_folder, 'split_base')
writer.dump()

