Partition¶
Error
This treatment is provided by an external module and is not part of the core system. Due to licensing restrictions, it is available exclusively for internal use at CERFACS
Description¶
Partition a mesh with the Python API of Gmsh.
Parameters¶
- source:
Baseor file path The input mesh to be partitioned. If the mesh has already been read by antares, provide the antares base. Otherwise, give the path to mesh file. In this case, the file must be able to be read by the Gmsh API.
- source:
- nb_parts: int, default= table below
Number of partitions to create. Depending on the execution mode (serial or MPI parallel) and the number of partitions demanded (nb_parts), the actual number of partitions that will be created is described in the table below.
nb_parts
serial
N process
None
1
N
M
M
error
- all_groups: bool, default= False
If True, all the physical groups in Gmsh are transferred to antares as zones. If False, only those physical groups are read into antares that have dimension equal to the model dimension. As an example, consider a 3D mesh with two physical groups. One is subset of the full 3D mesh, in which we expect that something interesting will happen. The other is a set of 2D faces where the boundary conditions are prescribed. If all_groups == False, the boundary surface meshes are not imported in antares. The reason this option exists is that some treatments in antares assume that the zones have equal dimensions, matching the dimension of the model.
- ghost: bool, default= False
If True, ghost elements are determined. Othewise, only partition-local elements.
Preconditions¶
gmsh and mpi4py are required dependencies.
Postconditions¶
If executed in serial, the output contains as many bases as the number of partitions. In parallel, a single base is returned on each process.
If ghost == True, ghost elements are stored in the 'ghost' attribute of the 'default'
zone. This attribute is a dictionary, storing the ghost elements as
{neighboring_partition_number: ghost_elements_on_that_partition}.
Example¶
The following code partitions mesh into 3 pieces.
import antares
myt = antares.Treatment('partition')
myt['source'] = 'delta.1.msh' # mesh shipped with antares
myt['nb_parts'] = 3 # do not define when running in parallel
myt['all_groups'] = True
partitioned_base = myt.execute()
Main functions¶
- class antares.plugins.cerfacs.treatment.TreatmentPartition.TreatmentPartition¶
Process to perform a Partition treatment with the Gmsh API.
- execute() Union[Base, List[Base]]¶
Call the partitioner.
- Returns:
antares base objects, one Base for every partition.
- Return type:
- Raises:
ValueError – If the partition count and the number of processes are not consistent.
FileNotFoundError – If the file to be partitioned does not exist.
TypeError – If the
sourceinput is not of the expected type.
- partition() None¶
Partition the mesh using Gmsh.
- See also:
Partitioning options are documented in the Mesh options section of the documentation.
- antares.plugins.cerfacs.treatment.TreatmentPartition.distribute(bases: List[Base]) Base¶
Distribute antares partitions among MPI processes.
The mesh partitioning in Gmsh is serial. When we want to work with a distributed mesh, the mesh partitions must be sent to the individual processes.
- Parameters:
bases (List[Base]) – antares base objects, one Base for every partition.
- Returns:
A single base object for every process.
- Return type:
- Notes:
Currently, antares bases are sent via MPI by first serializing them. While the serialization happens in memory (no file I/O), it is suboptimal: i) serialization is done with the pickle module of Python, ii) the complete base is duplicated. A better solution is to directly send the NumPy arrays that constitute the antares base. See the mpi4py tutorial.
- antares.plugins.cerfacs.treatment.TreatmentPartition.plot_mesh(mesh: List[Zone], with_ghosts: Optional[bool] = False, show_elem_labels: Optional[bool] = False, show_node_labels: Optional[bool] = False, colors: Optional[List] = None, separately: Optional[bool] = None) None¶
- Parameters:
mesh (List[Zone]) – Partitioned mesh, each partition is an antares zone
with_ghosts (Optional[bool]) – Whether to plot the ghost elements, defaults to False
show_elem_labels (Optional[bool]) – Whether to plot the element labels, defaults to False. This is a costly operation for large meshes because i) the element labels are additional text objects, ii) the centroid of each element must be computed to determine where to place the element label
show_node_labels (Optional[bool]) – Whether to plot the node labels, defaults to False.
colors (List[matplotlib.typing.ColorType], i.e. a color format accepted by matplotlib, optional) – Colors of the mesh partitions, one color for each partition
separately (Optional[bool]) – Whether to plot each mesh partition in a separate figure. If not given, and
with_ghosts is True, separate plots are created; ifwith_ghosts is False, all the mesh partitions are plotted in the same figure.
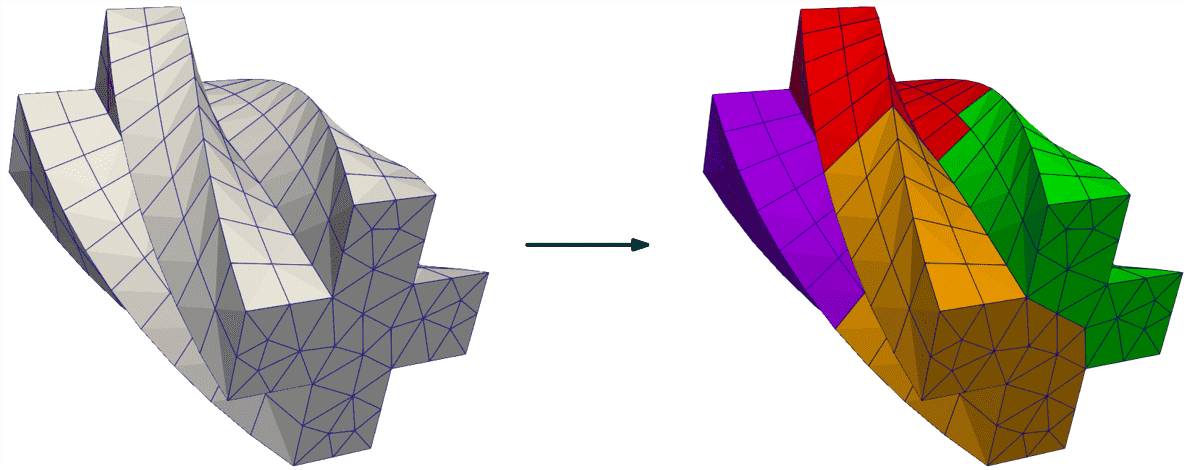
Example 1: Partition base and dump result¶
import os
import pathlib
from mpi4py import MPI
import antares
from antares.treatment.TreatmentPartition import plot_mesh
if not os.path.isdir('OUTPUT'):
os.makedirs('OUTPUT')
# Partition the mesh given in a file
TESTFILE = 'delta.1.msh' # feel free to experiment with other meshes and file types
path = pathlib.Path(__file__).parent.parent.parent / 'data' / 'GMSH' / TESTFILE
treatment = antares.Treatment('partition')
treatment['source'] = path
treatment['nb_parts'] = 3
treatment['all_groups'] = True
result = treatment.execute()
# Write the partitioned mesh to individual files, supporting both serial and parallel execution
if isinstance(result, list): # serial mode
index_base_mapping = [{i: part} for i, part in enumerate(result)]
else:
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
index_base_mapping = [{rank: result}]
# Dump each base
for mapping in index_base_mapping:
w = antares.Writer('hdf_antares') # the HDF output format is chosen because it can export the groups too
partition_index = list(mapping.keys())[0] # in parallel: MPI rank, in serial: list index
partition = list(mapping.values())[0]
w['base'] = partition
w['filename'] = os.path.join('OUTPUT', f'{path.stem}_{partition_index}.cgns')
w.dump()
Output of the first example:
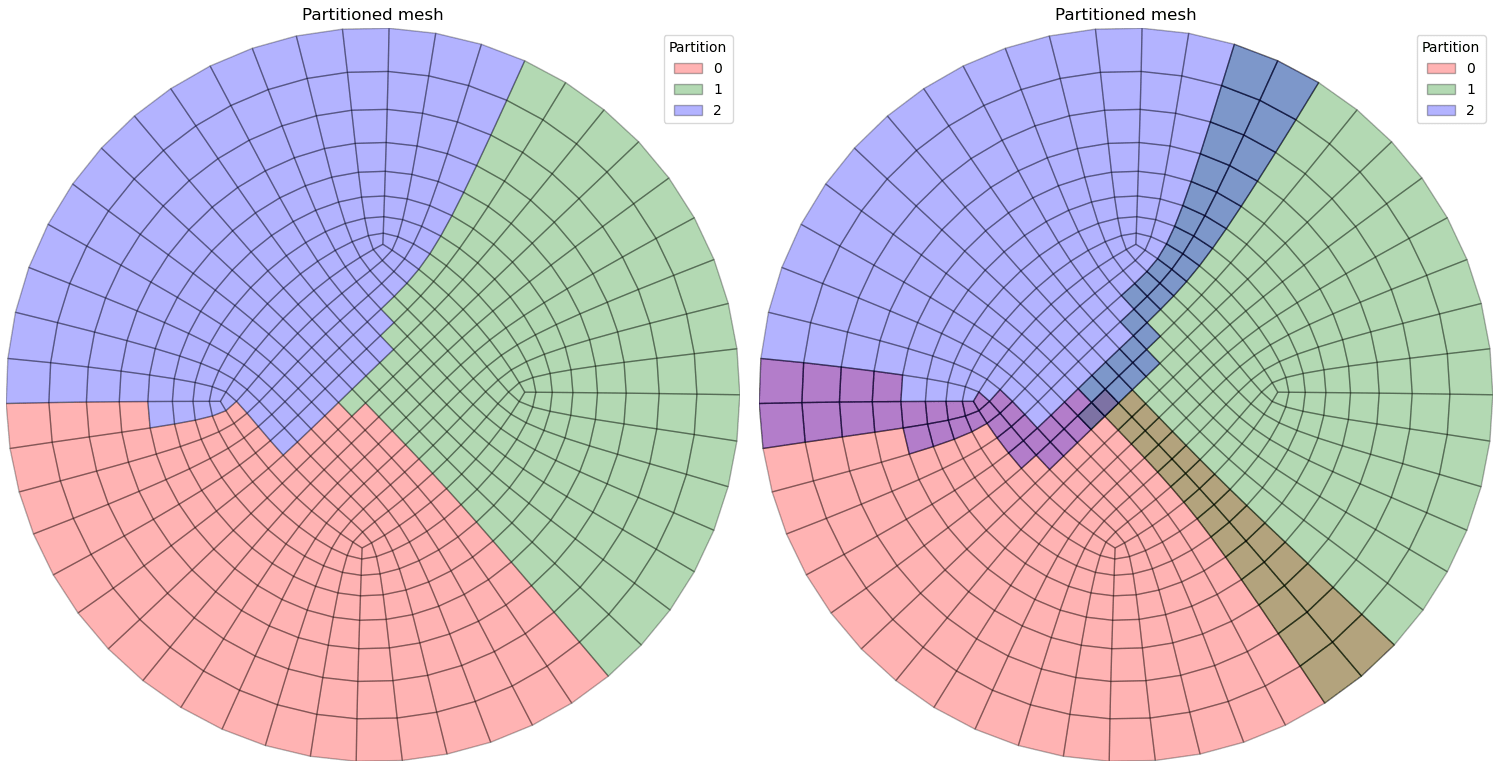
Example 2: Partition base and compute ghosts¶
import os
import pathlib
from mpi4py import MPI
import antares
from antares.treatment.TreatmentPartition import plot_mesh
if not os.path.isdir('OUTPUT'):
os.makedirs('OUTPUT')
TESTFILE = '2d.msh'
path = pathlib.Path(__file__).parent.parent.parent / 'data' / 'GMSH' / TESTFILE
treatment = antares.Treatment('partition')
treatment['source'] = path
treatment['nb_parts'] = 3
treatment['ghost'] = True
bases = treatment.execute()
# Plot partitions
plot_mesh([base['default'] for base in bases], with_ghosts=False, colors=['r', 'g', 'b'])
plot_mesh([base['default'] for base in bases], with_ghosts=True, colors=['r', 'g', 'b'])