Decimate¶
Description¶
Decimation of a structured or 2D unstructured grid using a reduction target.
This treatment can be used to coarsen a mesh containing a large number of elements.
The decimation process of 2D unstructured grids uses the VTK library to reduce the number of triangles.
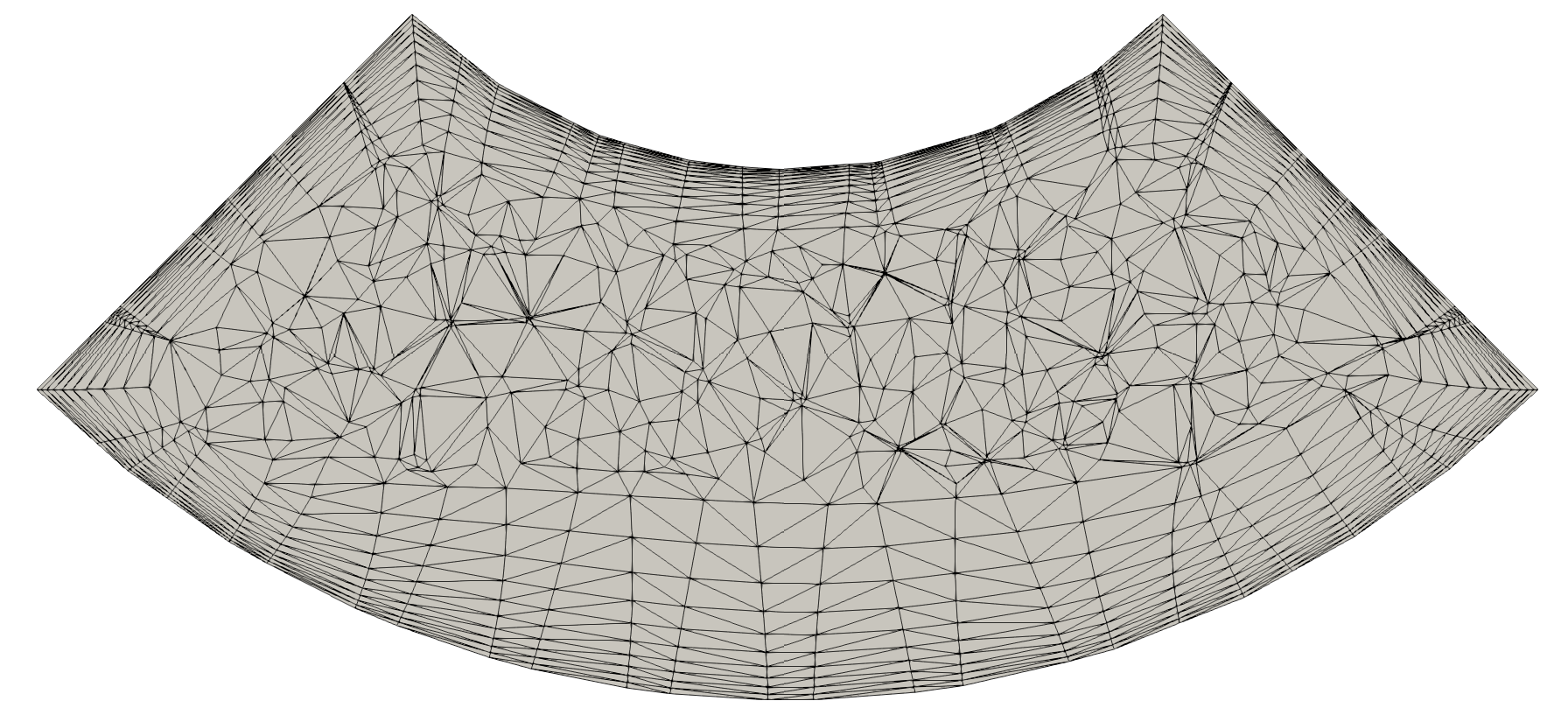
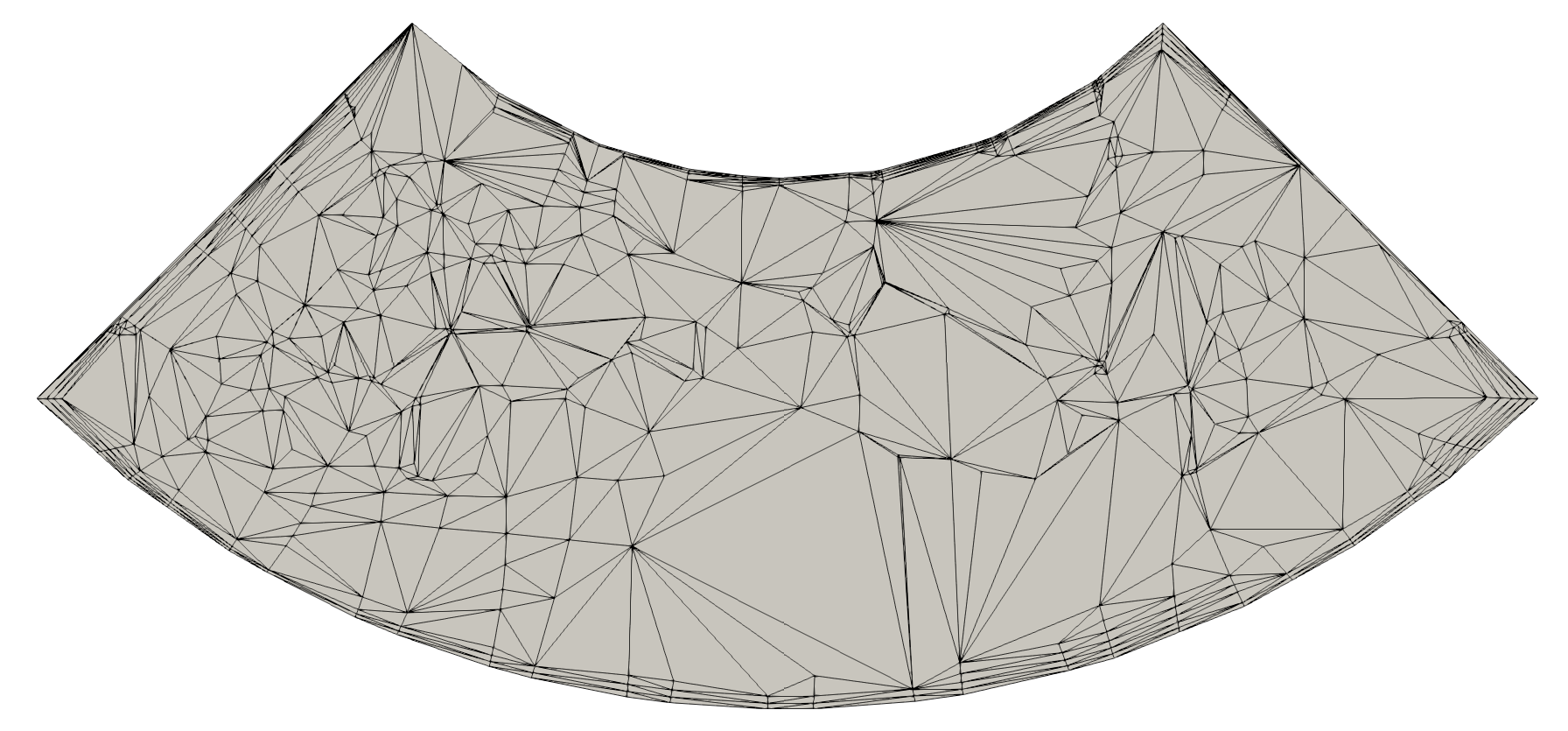
On the left: initial 2D unstructured (top) and structured (bottom) meshes.
On the right: resulting meshes after decimation with reduction = 0.5 for the unstructured mesh and reduction = 2. for the structured mesh.
Parameters¶
- base:
Base Base to decimate.
- base:
- coordinates: sequence(str)
The name of coordinates.
- reduction: float in [0,1] or int > 1,
For unstructured grids (2D only), the factor of reduction (float) specifies the percentage of triangles to be removed. For structured grids, the factor of reduction (integer) is the number of points to remove in each topological direction. Example: If the mesh contains 100 elements, and the reduction factor is 0.75, then after the decimation there will be approximately 25 elements - a 75% reduction
- memory_mode: bool, default= False
If True, the initial base is deleted on the fly to limit memory usage.
Preconditions¶
Zones can be either structured or 2D unstructured.
Zones can contain multiple instants.
Postconditions¶
The boundaries and families are not preserved in the output base.
Example¶
This example shows how to decimate a base.
import antares
myt = Treatment('decimate')
myt['base'] = base
myt['coordinates'] = ['x', 'y', 'z']
myt['reduction'] = 0.5
myt['memory_mode'] = True
decimated_base = myt.execute()
Main functions¶
Example¶
"""
This example shows how to decimate a base.
This can be useful if you want to test some kinds of treatment
on large meshes.
"""
import os
if not os.path.isdir('OUTPUT'):
os.makedirs('OUTPUT')
from antares import Reader, Treatment, Writer
# ------------------
# Reading the files
# ------------------
r = Reader('hdf_avbp')
r['filename'] = os.path.join('..', 'data', 'SECTOR', 'hybrid_sector.mesh.h5')
r['shared'] = True
base = r.read()
r = Reader('hdf_avbp')
r['base'] = base
r['filename'] = os.path.join('..', 'data', 'SECTOR', 'hybrid_sector.sol.h5')
r.read()
treatment = Treatment('cut')
treatment['base'] = base
treatment['type'] = 'plane'
treatment['origin'] = [0.0002, 0.0006, 0.]
treatment['normal'] = [1., 0., 0.]
result = treatment.execute()
ini_size = result[0][0].connectivity['tri'].size
# ------------------------------------------------------
# Decimate the unstructured base with a reduction factor
# ------------------------------------------------------
treatment = Treatment('decimate')
treatment['base'] = result
treatment['reduction'] = 0.1
treatment['memory_mode'] = True
result = treatment.execute()
print("Effective reduction: ", (100.0*(ini_size-result[0][0].connectivity['tri'].size))/ini_size, "%")
# -------------------
# Writing the result
# -------------------
w = Writer('bin_tp')
w['filename'] = os.path.join('OUTPUT', 'ex_uns_decimated.plt')
w['base'] = result
w.dump()
# ------------------
# Reading the files
# ------------------
reader = Reader('bin_tp')
reader['filename'] = os.path.join('..', 'data', 'ROTOR37', 'GENERIC', 'flow_<zone>_ite<instant>.dat')
base = reader.read()
# ------------------------------------------------------
# Decimate the unstructured base with a reduction factor
# ------------------------------------------------------
treatment = Treatment('decimate')
treatment['base'] = base
treatment['reduction'] = 2
treatment['memory_mode'] = True
result = treatment.execute()
# -------------------
# Writing the result
# -------------------
w = Writer('bin_tp')
w['filename'] = os.path.join('OUTPUT', 'ex_str_decimated.plt')
w['base'] = result
w.dump()