# Reading boundary data in a HDF-CGNS file¶
This tutorial shows how to:
read a turbomachinery configuration stored in HDF-CGNS format
compute the h/H variable
get the base corresponding to the blade given as a family
perform node-to-cell and cell-to-node on this latter base
make profile at some heights of the blade
output all curves in a single HDF-CGNS file
## Reading data
If we have a mesh file mesh.cgns and a solution file elsAoutput.cgns, then we can use the Antares reader hdf_cgns.
reader = antares.Reader('hdf_cgns')
reader['filename'] = 'mesh.cgns'
reader['shared'] = True
base = reader.read()
We put the mesh as shared variables.
reader = antares.Reader('hdf_cgns')
reader['base'] = base
reader['filename'] = 'elsAoutput.cgns'
base = reader.read()
We append the solution to the previous base.
## Computing h/H variable
The letter h means the hub, and the letter H the shroud. The h/H variable is the distance of a point from the hub on a line going from the hub to the shroud.
tr = antares.Treatment('hH')
tr['base'] = base
tr['row_name'] = 'ROW'
tr['extension'] = 0.1
tr['coordinates'] = ['CoordinateX', 'CoordinateY', 'CoordinateZ']
base = tr.execute()
The option ‘row_name’ tells the convention used to prefix the names of rows in the configuration. The option ‘extension’ is 10% of the radius of the configuration.
## Get the family base
Next, we extract the blade given by the family ROW(1)_BLADE(1)
row_1_blade_1 = base[base.families['ROW(1)_BLADE(1)']]
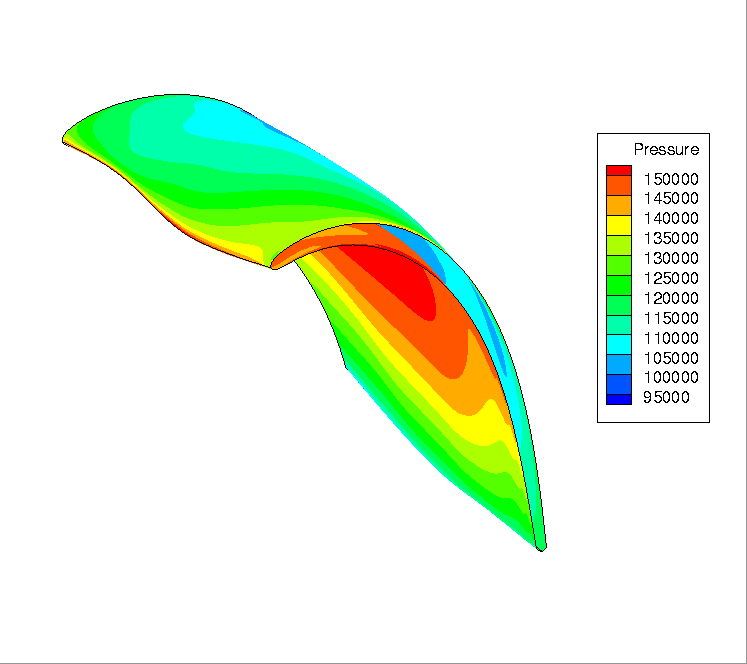
The pressure is stored in BC_t node of the HDF-CGNS file.¶
## Perform node-to-cell and cell-to-node on this latter base
The base row_1_blade_1 is a 2D base on which we can perform node2cell and cell2node operations.
row_1_blade_1.node_to_cell(variables=['ro'])
row_1_blade_1.cell_to_node(variables=['Pressure', 'Temperature'])
Then, we save the base in a HDF-CGNS file.
writer = antares.Writer('hdf_cgns')
writer['base'] = row_1_blade_1
writer['filename'] = 'all_wall_row_1_blade_1.cgns'
writer['base_name'] = 'row_1_blade_1'
writer['dtype'] = 'float32'
writer.dump()
Do not forget to remove the following file due to the append writing
try:
os.remove('all_iso.cgns')
except OSError:
pass
## Make profile at some heights of the blade ## Output all curves in a single HDF-CGNS file
Then, we loop on 5 heights.
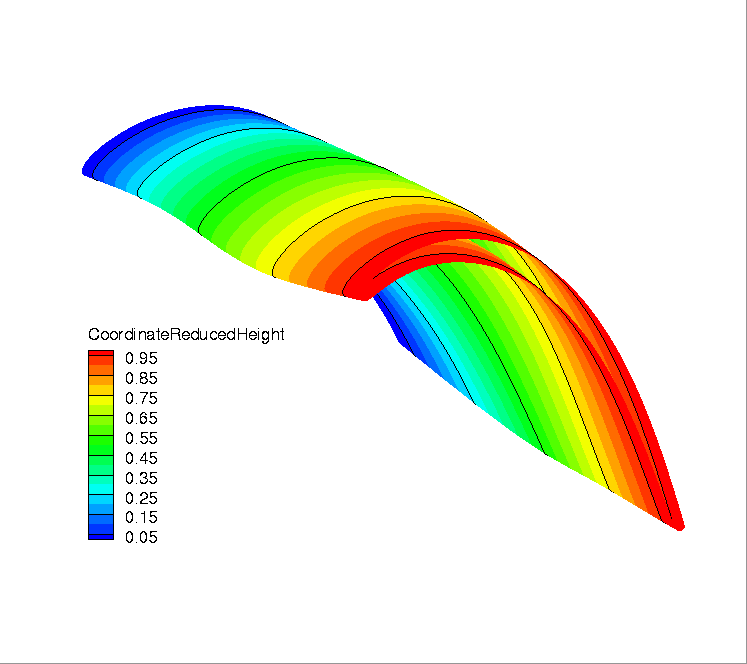
Location of heights on the blade.¶
for hoH_val in [0.05, 0.25, 0.50, 0.75, 0.95]:
t = antares.Treatment('isosurface')
t['base'] = row_1_blade_1
t['variable'] = 'CoordinateReducedHeight'
t['value'] = hoH_val
result = t.execute()
writer = antares.Writer('hdf_cgns')
writer['base'] = result
writer['filename'] = os.path.join('all_iso.cgns')
writer['append'] = True
writer['base_name'] = '%s' % hoH_val
writer['dtype'] = 'float32'
writer.dump()
‘CoordinateReducedHeight’ corresponds to the h/H variable. Note the option ‘append’ of the writer to concatenate results in a single file.
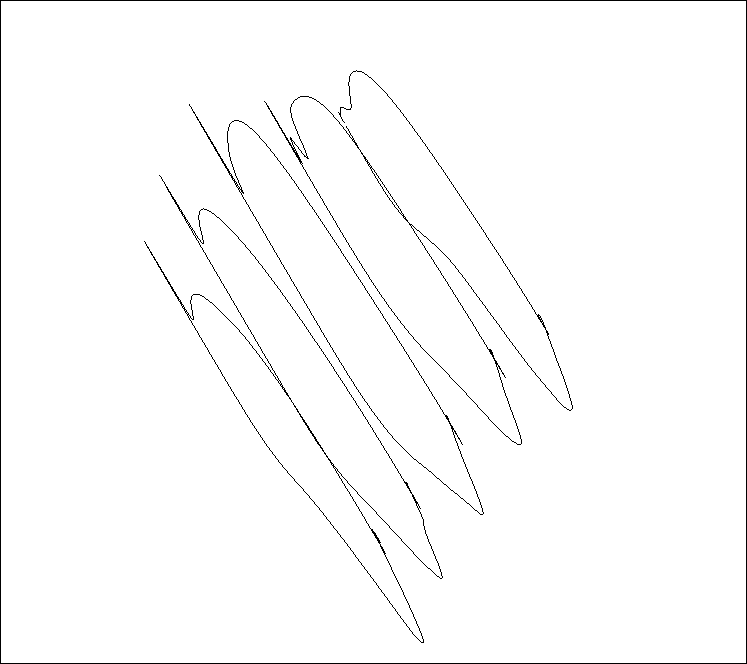
Pressure plots on the five blade profiles.¶
