CrinkleSlice¶
Description¶
Keep cells with the given value.
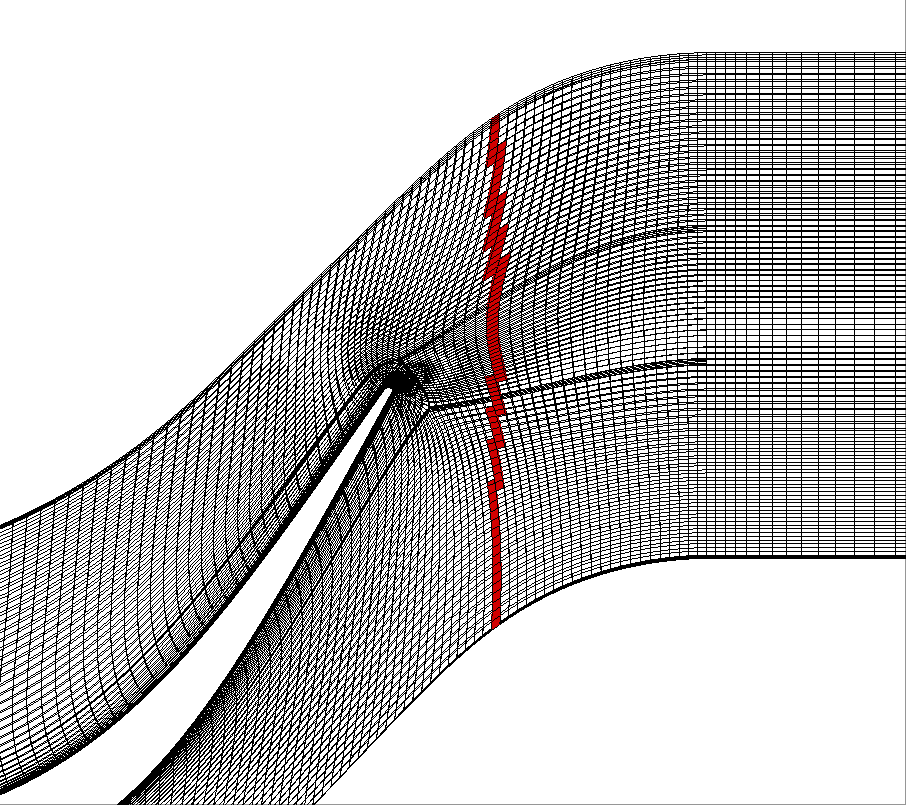
Initial grid in black and crinkle slice for x=70 in red.
Construction¶
import antares
myt = antares.Treatment('crinkleslice')
Parameters¶
- base:
Base The input base
- base:
- variable: str, default= None
The name of the variable used to extract cells.
- value: float
Keep cells with the given value.
- memory_mode: bool, default= False
If True, the initial base is deleted on the fly to limit memory usage
- with_families: bool, default= False
If True, the output of the treatment contains rebuilt families based on the input base. All the families and all the levels of sub-families are rebuilt, but only attributes and Zone are transfered (not yet implemented for Boundary and Window).
Preconditions¶
Postconditions¶
The output base is always unstructured. It does not contain any boundary condition.
The input base is made unstructured.
If with_families is enabled, the output base contains the reconstructed families of base.
Example¶
import antares
myt = antares.Treatment('crinkleslice')
myt['base'] = base
myt['variable'] = 'x'
myt['value'] = 70.
crinkle = myt.execute()
Main functions¶
Example¶
"""
This example shows how to extract a crinkle slice.
"""
import os
if not os.path.isdir('OUTPUT'):
os.makedirs('OUTPUT')
from antares import Reader, Treatment, Writer
# ------------------
# Reading the files
# ------------------
reader = Reader('bin_tp')
reader['filename'] = os.path.join('..', 'data', 'ROTOR37', 'GENERIC', 'flow_<zone>_ite0.dat')
ini_base = reader.read()
# -------------------------
# Crinkle Slice
# -------------------------
treatment = Treatment('crinkleslice')
treatment['base'] = ini_base
treatment['variable'] = 'x'
treatment['value'] = 70.
result = treatment.execute()
# -------------------
# Writing the result
# -------------------
writer = Writer('hdf_antares')
writer['filename'] = os.path.join('OUTPUT', 'ex_crinkleslice')
writer['base'] = result
writer.dump()